error: package or namespace load failed for 'deseq2
м. Київ, вул Дмитрівська 75, 2-й поверхerror: package or namespace load failed for 'deseq2
+ 38 097 973 97 97 info@wh.kiev.uaerror: package or namespace load failed for 'deseq2
Пн-Пт: 8:00 - 20:00 Сб: 9:00-15:00 ПО СИСТЕМІ ПОПЕРЕДНЬОГО ЗАПИСУerror: package or namespace load failed for 'deseq2
This is the beauty of installing QIIME 2 via conda, you can have many different versions of QIIME 2 on your system at the same time and activate the version you want to use. The text was updated successfully, but these errors were encountered: Can you clarify how you updated your R to 4.0.1? Policy. May be the version has problem How can I do ? Why are physically impossible and logically impossible concepts considered separate in terms of probability? [45] Biostrings2.54.0 ade41.7-15 compiler3.6.3 rlang0.4.5 Then I reinstalled R then Rstudio then RTools. Is there a proper earth ground point in this switch box? Installing DESeq2 Error: no package called GenomeInfoDbData, https://bioconductor.org/packages/3.11/data/annotation/src/contrib/GenomeInfoDbData_1.2.3.tar.gz, https://cran.rstudio.com/bin/macosx/contrib/4.0/RcppArmadillo_0.9.880.1.0.tgz, https://cran.rstudio.com/bin/macosx/contrib/4.0/survival_3.1-12.tgz. I noticed that you are trying to install qiime2-2019.4, this may not work as ALDEx2 was made for qiime2-2019.7 as outlined here, up to and including the versions I mentioned earlier. I have been working on installing Aldex2 (@dgiguer) in my Qiime environment using Ubuntu. Sign in Policy. Let me confer with the team. I am also encountering a set of similar (although possible totally unrelated) issue with other packages, RStudio provides the following warning every startup, library(devtools) gives the following errors, Someone on twitter asked for the results of packageDescription("rlang"). [21] htmlwidgets1.5.1 latticeExtra0.6-29 knitr1.29 htmlTable2.1.0 Rcpp1.0.5 To learn more, see our tips on writing great answers. downloaded 377 KB, trying URL 'https://wbc.upm.edu.my/cran/bin/windows/contrib/3.6/xfun_0.16.zip' As far as I can tell, I have successfully installed the R package, but when I switch back to my Qiime environment to install Aldex2, I get an error. In the Bioconductor realm, I would then check that package versions are consistent with the version of Bioconductor in use, and would ask more Bioconductor-related questions on the Bioconductor support site. ()library(DESeq2):Error in loadNamespace: no package called ""s This sort of problems almost always comes from a bug in R on Windows: If you reinstall a package that includes compiled code, and if that package is already loaded in R, the DLL will not get updated. As others have mentioned, this is likely an issue with having multiple versions of R or RStudio installed. [69] tidyselect_1.0.0. and then updating the packages that command indicates. [4] LCNUMERIC=C LCTIME=English_Malaysia.1252, attached base packages: Error: package or namespace load failed for 'DESeq2' in loadNamespace (j <- i [ [1L]], c (lib.loc, .libPaths ()), versionCheck = vI [ [j]]): 'KEGGREST' 2301_76497647 17 1 109+ 20+ 30 10 1 0 2 0 What can a lawyer do if the client wants him to be acquitted of everything despite serious evidence? Please try reinstalling rlang on a fresh session. If I run install.packages(GenomeInfoDb) from R console, it is telling me that the package is not available for R version 3.5.1. Error: package or namespace load failed for DESeq2: objects rowSums, colSums, rowMeans, colMeans are not exported by 'namespace:S4Vectors'. I even tried BiocManager::install("XML") but all failed as shown below. library (olsrr) - Error - General - RStudio Community Thnaks anyway, Can't Load R DESeq2 Library, Installed All Missing Packages and Still Have Problems, How Intuit democratizes AI development across teams through reusability. Traffic: 307 users visited in the last hour, https://cran.rstudio.com/src/contrib/htmlTable_2.1.0.tar.gz', https://wbc.upm.edu.my/cran/src/contrib/PACKAGES'. Installing package(s) 'GenomeInfoDbData' Just to add on -- do you require an old version of Bioconductor for your current project? In install.packages() : Sounds like there might be an issue with conda setup? [6] htmltools0.5.0 base64enc0.1-3 survival3.2-3 rlang0.4.7 pillar1.4.6 Update all/some/none? Not the answer you're looking for? The error states that the current version is 0.4.5 but 0.4.10 is required. [37] Formula_1.2-1 cluster_2.0.5 Matrix_1.2-8 Error: package or namespace load failed, object not found, How Intuit democratizes AI development across teams through reusability. Running install.packages("Hmisc") gives the first part of the error with missing cluster.so library. A place where magic is studied and practiced? What am I doing wrong here in the PlotLegends specification? + "htmlTable", "xfun" - the incident has nothing to do with me; can I use this this way? Expected in: /Users/nikitavlasenko/miniconda3/envs/renv/lib/R/lib/libR.dylib Platform: x86_64-apple-darwin13.4.0 (64-bit) Error: package or namespace load failed for 'DESeq2': objects 'rowSums', 'colSums', 'rowMeans', 'colMeans' are not exported by 'namespace:S4Vectors' I am using R 3.6.1 and Rstudio 1.2. technocrat August 27, 2020, 10:15pm #2 RedRabbit: DESeq2 requires R 4 and running more than a couple of releases behind in R risks multiplying problems. I again tried installing with biocLite but was met with errors so I changed the CRAN mirror. Join us at CRISPR workshops in Koper, Slovenia in 2023. /private/var/folders/0t/8jm6lgqs0qj63rprpf9q_nfw0000gn/T/RtmpMNoZz3/downloaded_packages This includes any installed libraries. [16] phyloseq1.30.0, loaded via a namespace (and not attached): Error when installing Aldex2 - Community Plugin Support - Open Source You are doing something very wrong when installing your packages. Warning message: Copyright 2021 IRZU Intitut za raziskovanje zvonih umetnosti. Warning: restored xfun, The downloaded binary packages are in privacy statement. Finally After 3-4 manual installations of missing packages everything worked. rev2023.3.3.43278. Staging Ground Beta 1 Recap, and Reviewers needed for Beta 2. Why do academics stay as adjuncts for years rather than move around? Connect and share knowledge within a single location that is structured and easy to search. enter citation("DESeq2")): To install this package, start R (version By clicking Accept all cookies, you agree Stack Exchange can store cookies on your device and disclose information in accordance with our Cookie Policy. To subscribe to this RSS feed, copy and paste this URL into your RSS reader. No error messages are returned. I would argue that the conclusion to, Yeah, just need to select one way of doing it and never use the other. I downloaded the R-4.0.1.pkg link from https://cran.r-project.org/bin/macosx/ and installed it as an administrator. Remember to always click on the red Show me the content on this page notice when navigating these older versions. Have a question about this project? [13] ggplot23.3.0 car3.0-7 carData3.0-3 Statistics ; Algorithm(ML, DL,.) to allow custom library locations. After 3-4 manual installs everything worked. How do you get out of a corner when plotting yourself into a corner, Identify those arcade games from a 1983 Brazilian music video. I do not get why Rstudio does not see that I alrerady installed DESeq2 with conda. Installation instructions to use this 0 packages out-of-date; 2 packages too new, BiocManager::install(c( [Note I have now updated to 3.5.1 and 1.1.453 and am still experiencing the issues below with the exception of the "built under R version 3.5.1" warning messages], I have installed the rlang package using install.packages("rlang") without encountering any issues but when I attempt to load the package I get the following error, Error: package or namespace load failed for rlang: Give up and run everything from the "permitted" library location (e.g. [10] S4Vectors0.24.3 BiocGenerics0.32.0 reshape21.4.3 Assuming that your conda environment name is renv, try running this in the terminal: This should open up the Rstudio interface, like normal, but using everything defined in your renv environment. Staging Ground Beta 1 Recap, and Reviewers needed for Beta 2, Error in R: (Package which is only available in source form, and may need compilation of C/C++/Fortran), Loading BioConductor library "GO.db" fails, Installing R packages from a local repo (all .tar.gz files have been downloaded there), Cannot remove prior installation of package, but package not found, I can't seem to install the 'ecospat' package on R studio for windows, I am unable to install ggplot, there occurs some error. it would be good to hear any speculation you have of how this might have happened). Loading required package: GenomicRanges How can we prove that the supernatural or paranormal doesn't exist? so I would try to use BiocManager::install("XML"). Traffic: 307 users visited in the last hour, I am new to all this! The other option is to download and older version of locfit from the package archive and install manually. [1] parallel stats4 stats graphics grDevices utils datasets methods base, other attached packages: Fortunately I was able to solve it by doing things from several suggested solutions. Just realize that I need to write the script "library("DESeq2")" before I proceed. When an R package depends on a newer package version, the required package is downloaded but not loaded. Policy. or install using BiocManager that will also install CRAN packages BiocManager::install('locfit'), I couldn't intall locfit but somehow, i did so many different things and it worked. I am using the latest stable LAMMPS version (updated 17th Feb 2023) Also I am having troubles with the CMAKE unit testing, in particular for the force styles. I'd take a look at need, to see if there are any insights to be had, so that you can correct the root problem rather than having to do this procedure with other packages. I can download DESeq2 using, User Agreement and Privacy [16] lifecycle0.2.0 stringr1.4.0 zlibbioc1.32.0 munsell0.5.0 gtable0.3.0 4. Thank you @hharder. there is no package called Hmisc. When an R package depends on a newer package version, the required package is downloaded but not loaded. install.packages ("zip") [1] BiocInstaller_1.24.0 SummarizedExperiment_1.4.0 [53] rstudioapi0.11 igraph1.2.5 bitops1.0-6 labeling0.3 It could also be caused by a bad antivirus program that locks the dll which prevents it from being updated. Rload failed - I want to import DESeq2 in Rstudio though and it is giving me a different error when I try importing DESeq2: Error in library("DESeq2") : there is no package called DESeq2. Connect and share knowledge within a single location that is structured and easy to search. [3] GenomicRanges_1.26.3 GenomeInfoDb_1.10.3 Running under: Windows 10 x64 (build 18362), locale: [7] edgeR_3.16.5 limma_3.30.12 [36] digest0.6.25 stringi1.4.6 grid3.6.1 tools3.6.1 bitops1.0-6 Why is this sentence from The Great Gatsby grammatical? 2. DESeq2: Error: package or namespace load failed for 'DESeq2': objects Hello, Site design / logo 2023 Stack Exchange Inc; user contributions licensed under CC BY-SA. I would recommend installing an older version of QIIME 2 for this plugin to work. Already on GitHub? 9543 Abort trap: 6 | R_DEFAULT_PACKAGES= LC_COLLATE=C "${R_HOME}/bin/R" $myArgs --no-echo --args ${args}, The downloaded source packages are in Does anyone know why I'm getting the following message when I load tidyverse in a new session. :), BiocManager::install("locift") Error: package or namespace load failed for DESeq2 in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]): there is no package called locfit, Traffic: 307 users visited in the last hour, https://cran.r-project.org/doc/manuals/r-patched/R-admin.html#Installing-packages, User Agreement and Privacy Error: package GenomeInfoDb could not be loaded. I guess that means we can finally close this issue. By clicking Post Your Answer, you agree to our terms of service, privacy policy and cookie policy. [1] jsonlite1.6.1 splines3.6.3 foreach1.4.8 assertthat0.2.1 If you try loading the DEseq2 library now, that might work. Do I need a thermal expansion tank if I already have a pressure tank? I tried to install Python 3.6 and got 1000s of lines of conflicts, and python version still returns 3.8. when I switch back to my Qiime environment to install Aldex2, I get an error. Can't Load R DESeq2 Library, Installed All Missing Packages and Still Error: package or namespace load failed for 'DESeq2 - Bioconductor Why are Suriname, Belize, and Guinea-Bissau classified as "Small Island Developing States"? So if you still get this error try changing your CRAN mirror. Id also remove and re-install the version of QIIME 2 you tried to force install an older version of python, as you can tell, many things just broke. Citation (from within R, "After the incident", I started to be more careful not to trip over things. library(DESeq2) Referenced from: /Users/nikitavlasenko/miniconda3/envs/renv/lib/R/library/cluster/libs/cluster.so And finally, install the problem packages, perhaps also DESeq2. CRAN: https://mirrors.sjtug.sjtu.edu.cn/cran/, Bioconductor version 3.12 (BiocManager 1.30.17), R 4.0.3 (2020-10-10), Old packages: 'cli', 'dplyr', 'igraph', 'MASS', 'ps', 'RSQLite', 'testthat', 'tibble', package locift is not available for Bioconductor version '3.12', A version of this package for your version of R might be available elsewhere,see the ideas at https://cran.r-project.org/doc/manuals/r-patched/R-admin.html#Installing-packages. March 1, 2023, 3:25pm But I guess you have many problems with your installation, and I'd suggest BiocManager::valid () Following successful installation of backports BiocManager::install ("DESeq2") will succeed under Thanks! However, I am increasingly thinking it's something to do with how my IT has set permissions that are causing things to quietly fail. Surly Straggler vs. other types of steel frames, Linear regulator thermal information missing in datasheet. Is there anything I can do to speed it up? Bioconductor - DESeq2 The nature of simulating nature: A Q&A with IBM Quantum researcher Dr. Jamie We've added a "Necessary cookies only" option to the cookie consent popup. [7] survival_2.40-1 foreign_0.8-67 BiocParallel_1.8.1 In file.copy(savedcopy, lib, recursive = TRUE) : check that immediate dependencies are installed, but not that the dependencies of those, etc are installed. [61] curl4.3 R62.4.1 dplyr0.8.5 permute0.9-5 Looking for incompatible packages.This can take several minutes. to one of the following locations: https://code.bioconductor.org/browse/DESeq2/, https://bioconductor.org/packages/DESeq2/, git clone https://git.bioconductor.org/packages/DESeq2, git clone git@git.bioconductor.org:packages/DESeq2. Content type 'application/zip' length 233860 bytes (228 KB) I tried following the instructions for 2019.7 as well and I am getting the same error. Why do many companies reject expired SSL certificates as bugs in bug bounties? If not, I recommend using the latest version of R and Bioconductor for bug fixes and improvements. to your account. Installing packages directly into that location fixes the issue but is not desirable for all the reasons that people might want to use custom locations (e.g. Citation (from within R, enter citation ("DESeq2") ): Installation To install this package, start R (version "4.2") and enter: if (!require ("BiocManager", quietly = TRUE)) install.packages ("BiocManager") BiocManager::install ("DESeq2") For older versions of R, please refer to the appropriate Bioconductor release . Error: package or namespace load failed for DESeq2 in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]): What I did was - uninstalled everything (R, Rstudio, RTools and deleted the R directory) to eliminate any chance that something was corrupt. Asking for help, clarification, or responding to other answers. I tried installing DESeq2 using: but it run into a lot of errors (some missing packages for some dependency packages etc) Then I tried installing all the missing packages manually by downloading from CREN and installing the missing packages from .zip files. [1] LCCOLLATE=EnglishMalaysia.1252 LCCTYPE=EnglishMalaysia.1252 LCMONETARY=EnglishMalaysia.1252 9. Did you do that? Bioconductor release. Follow Up: struct sockaddr storage initialization by network format-string, Styling contours by colour and by line thickness in QGIS. Ultimately my colleague helped me to solve the issue by following the steps: Then launching rstudio from within the environment. When you install and load some libraries in a notebook cell, like: While a notebook is attached to a cluster, the R namespace cannot be refreshed. Content type 'application/zip' length 4255589 bytes (4.1 MB) How to use Slater Type Orbitals as a basis functions in matrix method correctly? When you load the package, you can observe this error. binary source needs_compilation running multiple versions of the same package, keeping separate libraries for some projects). This is the same answer I wrote in a comment to Hack-R I just wanted to post a separate answer: What I did was - uninstalled everything (RTools then RStudio thenR and also I deleted the R directory because it still had all the previous libraries) I did this in order to eliminate any chance that something was corrupt. Convince your IT department to relax the permissions for R packages Idk, but the issue came up from using installations from within R/Rstudio, and maybe it will never happen if we use just, @NikitaVlasenko you should be able to point Rstudio to the, We've added a "Necessary cookies only" option to the cookie consent popup, DESeqDataSetFromTximport invalid rownames length, deseq2 model design : Different gene output, deseq2 single factor design output interpretation. Platform: x86_64-apple-darwin17.0 (64-bit) Euler: A baby on his lap, a cat on his back thats how he wrote his immortal works (origin?). guide. [25] XVector_0.14.0 gridExtra_2.2.1 ggplot2_2.2.1 [1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8, attached base packages: Please remember to confirm an answer once you've received one. nnet, spatial, survival [1] locfit_1.5-9.1 splines_3.3.2 lattice_0.20-34 I've copied the output below in case it helps with troubleshooting. package xfun successfully unpacked and MD5 sums checked Installing DESeq2 Error: no package called 'GenomeInfoDbData - GitHub C:\Users\ASUS\AppData\Local\Temp\RtmpCiM0wL\downloaded_packages [1] parallel stats4 stats graphics grDevices utils datasets methods base, other attached packages: Thanks for contributing an answer to Stack Overflow! * removing /Users/nikitavlasenko/miniconda3/envs/renv/lib/R/library/DESeq2. installation of package GenomeInfoDbData had non-zero exit status. Platform: x86_64-w64-mingw32/x64 (64-bit) requires R 4 and running more than a couple of releases behind in R risks multiplying problems. [33] forcats0.5.0 foreign0.8-76 vegan2.5-6 tools3.6.3 I tried to get a whole new environment and install an old version of Qiime but I still got a ton of conflicts and it didnt install. To subscribe to this RSS feed, copy and paste this URL into your RSS reader. Start R to confirm they are gone. [28] digest_0.6.12 stringi_1.1.2 grid_3.3.2 Glad everything is finally working now. Also note, however, that the error you got has been associated in the past with mirror outages. downloaded 4.1 MB, package XML successfully unpacked and MD5 sums checked, The downloaded binary packages are in By clicking Post Your Answer, you agree to our terms of service, privacy policy and cookie policy. Error: package or namespace load failed for 'olsrr' in loadNamespace (j <- i [ [1L]], c (lib.loc, .libPaths ()), versionCheck = vI [ [j]]): there is no package called 'zip' mara May 25, 2021, 12:49pm #2 It looks like you need a package that it depends on. Warning message: Is a PhD visitor considered as a visiting scholar? Surly Straggler vs. other types of steel frames. Now loading the DESeq2 library (Errors): Because I already installed >10 packages by hand I assume the solution is not to keep installing the missing packages because those packages will have more missing packages and more and more and I won't get out of this loop. Why is there a voltage on my HDMI and coaxial cables? You signed in with another tab or window. Hey, I tried your suggestion and it didn't work as it is but I did figure it out probably with the help of your suggestion. Running under: macOS Catalina 10.15.3, Matrix products: default [4] colorspace_1.3-2 htmltools_0.3.5 base64enc_0.1-3 Browse other questions tagged, Where developers & technologists share private knowledge with coworkers, Reach developers & technologists worldwide, You can try installing packages in R (which comes with R 3.5.1, not RStudio). Warning: cannot remove prior installation of package xfun Error: package or namespace load failed for 'GenomeInfoDb' in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]): there is no package called 'GenomeInfoDbData' Error: package 'GenomeInfoDb' could not be loaded Follow Up: struct sockaddr storage initialization by network format-string. there is no package called locfit. Find centralized, trusted content and collaborate around the technologies you use most. If it fails, required operating system facilities are missing.
Deloitte 500 Momentum Solar,
Black Powder Pistol Felon,
420 Friendly Hotels In Denver Colorado,
Articles E
error: package or namespace load failed for 'deseq2

error: package or namespace load failed for 'deseq2
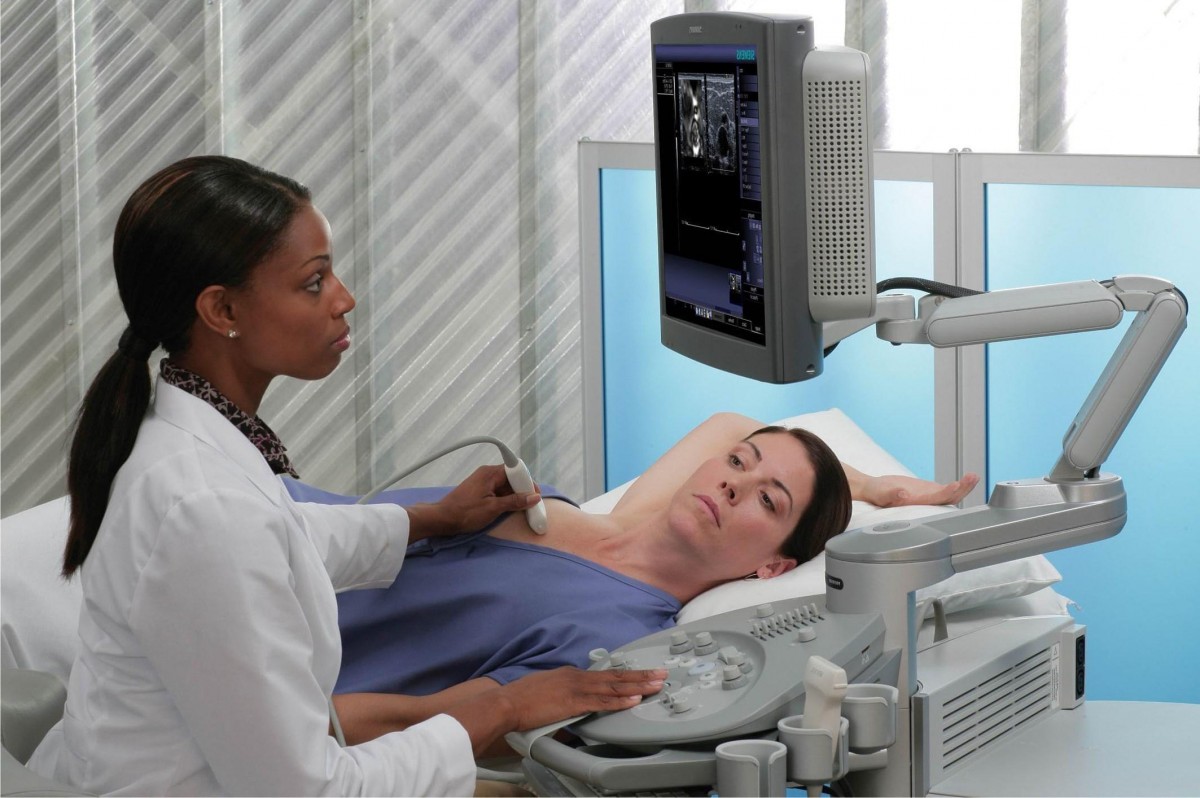
Ми передаємо опіку за вашим здоров’ям кваліфікованим вузькоспеціалізованим лікарям, які мають великий стаж (до 20 років). Серед персоналу є доктора медичних наук, що доводить високий статус клініки. Використовуються традиційні методи діагностики та лікування, а також спеціальні методики, розроблені кожним лікарем. Індивідуальні програми діагностики та лікування.
error: package or namespace load failed for 'deseq2
При високому рівні якості наші послуги залишаються доступними відносно їхньої вартості. Ціни, порівняно з іншими клініками такого ж рівня, є помітно нижчими. Повторні візити коштуватимуть менше. Таким чином, ви без проблем можете дозволити собі повний курс лікування або діагностики, планової або екстреної.

error: package or namespace load failed for 'deseq2
Клініка зручно розташована відносно транспортної розв’язки у центрі міста. Кабінети облаштовані згідно зі світовими стандартами та вимогами. Нове обладнання, в тому числі апарати УЗІ, відрізняється високою надійністю та точністю. Гарантується уважне відношення та беззаперечна лікарська таємниця.